The Epigenetics of Fish in Warming Water.
Many years ago, on a website where I was ultimately banned for telling the truth, I made fun of the "science" of Joseph Stalin:
The Most Interesting Chemistry of Lenin's Dead Body.
In this post about the "real stable genius" Stalin, and his relationship to Ilya Zbarsky and his father, I referred to Stalin's belief in "Lysenkoism," a belief which rejected natural selection and genetics and substituted a nonsense theory about heritability of acquired characteristics, a system of beliefs harking back to the ideas of Lamarck.
Stalin's faith based belief in the work of Lysenko set Soviet biological science back decades, resulting in the collapse, among other things, of Soviet grain harvests even years after Stalin had kicked off, much to the improvement of the world in general.
While it is true that natural selection is safe from political stupidity in most places - the rather communist style Republican Party notwithstanding - it is also true that a case can be made for genetic transmission of environmental conditions even without changes to DNA sequences, something.
This area, which has been burgeoning only in the last 20 years or so, is the fascinating subject of epigentics.
A very nice lecture on this topic, epigenetics is available on line: Professor Shirley Tilghman: The Wild and Wacky World of Epigenetics I attended this lecture when it was given and it was really, really informative and I recommend it highly.
Epigenetics involves the chemical transformation of the nuclear bases, often in the form of methylation of a cytosine, which is controlled by the proteins wrapping DNA, proteins known as histones, these in turn being controlled by post-translational modifications of the protein sequence by, for example, methylation or acetylation of lysine residues which are prominent in the sequences of histones.
Epigenetics, which also can involve the bonding of far more complex molecules, including many pollutants, is thought to result in many diseases, notably cancer, as well as somatic mutations. (One of my sons has a somatic mutation that resulted in a birth defect that proved to be minor though it need not have been. It involved, apparently, the deamination of a guanine moiety during gestation.)
In the most recent issue of Nature Climate Change epigenetic changes to fish are reported.
Some of these modifications are, despite the deserved rejection of Lysenko/Lamarckian ideas or ideology, heritable.
The paper is here: The epigenetic landscape of transgenerational acclimation to ocean warming (Munday et al Nature Climate Change Volume 8, Pages 504–509 (2018))
The introduction:
Epigenetic inheritance is a potential mechanism by which the environment in one generation can influence the performance of future generations1. Rapid climate change threatens the survival of many organisms; however, recent studies show that some species can adjust to climate-related stress when both parents and their offspring experience the same environmental change2,3. Whether such transgenerational acclimation could have an epigenetic basis is unknown. Here, by sequencing the liver genome, methylomes and transcriptomes of the coral reef fish, Acanthochromis polyacanthus, exposed to current day (+ 0 °C) or future ocean temperatures (+ 3 °C) for one generation, two generations and incrementally across generations, we identified 2,467 differentially methylated regions (DMRs) and 1,870 associated genes that respond to higher temperatures within and between generations. Of these genes, 193 were significantly correlated to the transgenerationally acclimating phenotypic trait, aerobic scope, with functions in insulin response, energy homeostasis, mitochondrial activity, oxygen consumption and angiogenesis. These genes may therefore play a key role in restoring performance across generations in fish exposed to increased temperatures associated with climate change...
Some elaboration:
Recently, we have shown that the common reef fish, Acanthochromis polyacanthus, can fully acclimate its scope for oxygen consumption (net aerobic scope) when both parents and their offspring experience the same increase in water temperature2,9. They do this by changing their transcriptional regulation of metabolism, cytoprotection, immunity, growth and cellular organization. Furthermore, these fish that are transgenerationally exposed to 3 °C warmer water (transgenerational treatment) differentially express a similar suite of genes compared with fish that are exposed to elevated temperatures from early development for just one generation (developmental treatment), albeit with more genes and higher magnitude changes in expression9. Reproductive capacity, however, was impaired in developmental and transgenerational fish, and only improved when temperature was increased incrementally (step-wise treatment) across two generations10. Here, we investigate if genomic DNA methylation could be implicated in the observed transgenerational plasticity of A. polyacanthus in an ocean warming scenario.
The methylation at the 5 position of a cytosine often initiates DNA repair, however under some conditions, the cytosine can deaminate to be converted into thymine, in which case a permanent mutation results.
I don't have a lot of time tonight, so I'll just cut to the pictures, which often is enough to induce an understanding of a paper if one hasn't much time.
The experimental set up:
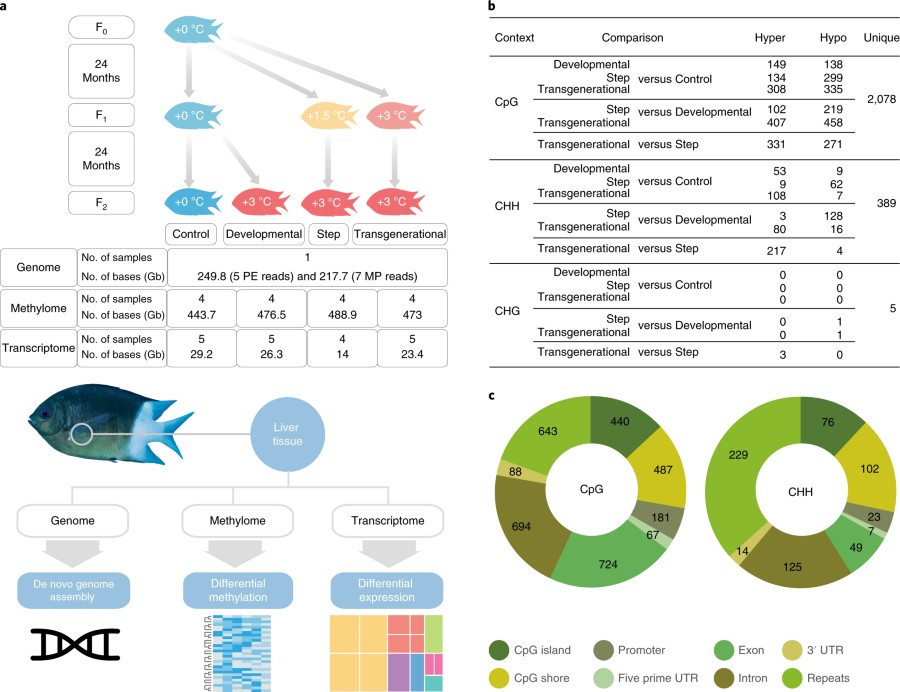
The caption:
Fig. 1 | Experimental design and summary of the DMRs. a, Design of the fish rearing experiment and summary statistics of the genome, methylomes and transcriptomes. b, The number of DMRs between treatments for three methylation contexts. Hyper and hypo indicate higher and lower methylation, respectively, in the treatment in the left of the Comparison column compared to the right. The Unique column represents the unique number of DMRs in each methylation context. c, DMR distribution across genomic elements for CpG and CHH contexts. The CHG context is not shown due to the low number of DMRs.
CpG is a cytosine phosphate guanine system. "CHH" or "CHG" refer to triples where H can be adenine, thymine or another cytosine.
Some plots of the methylation patterns of fish in different groups.
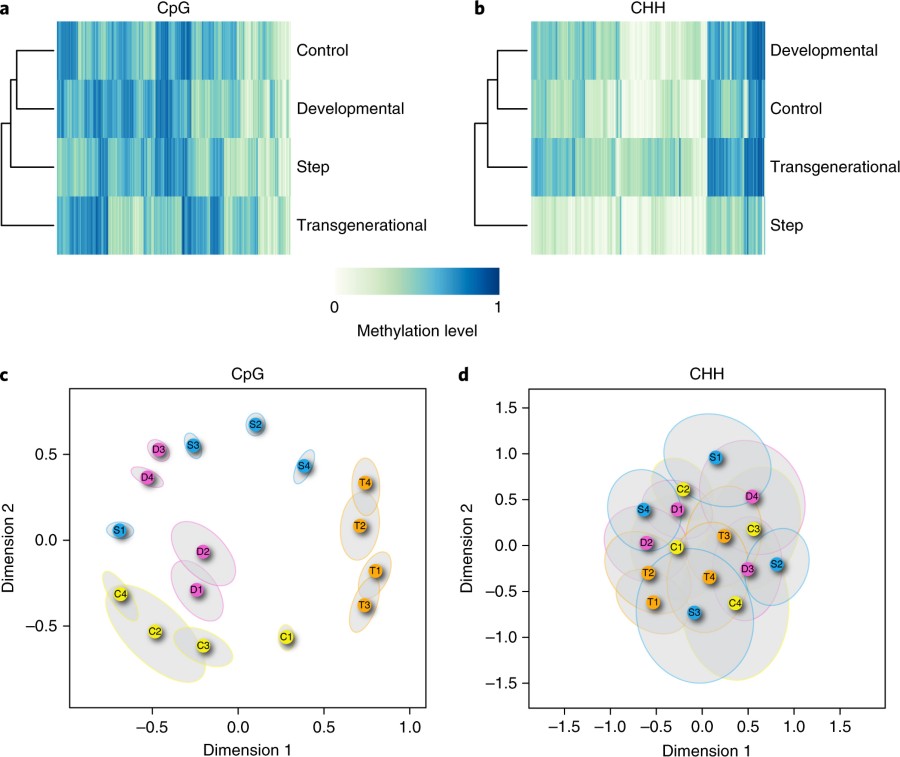
The caption:
Fig. 2 | Differential methylation patterns. a,b, Heatmap of DMRs for CpG (a) and CHH (b) contexts. c,d, MDS plot of DMRs for CpG (c) and CHH (d) contexts. Each coloured circle represents one fish sample and treatment groups are denoted by different colours. C, D, S and T represent control, developmental, step and transgenerational treatments, respectively. Each ellipse represents a 95% confidence region from 1,000 bootstrapping of DMRs from each sample. Non-overlapping ellipses implies statistically significant differences among samples.
A heatmap of the genes involved in respiration and their differential methylation:

The caption:
Fig. 3 | Heatmaps of differentially methylated and net aerobic scopecorrelated genes. a,b, Negatively (a) and positively (b) correlated differentially methylated genes (adjusted p < 0.05; > 25% difference in methylation between treatments) from A. polyacanthus, comparing control, developmental, transgenerational and step treatments. Genes described in the text are marked in bold. The colour scale indicates the per cent difference in methylation between two treatments
Some more results:

The caption:
Fig. 4 | DNA methylation patterns for thermal acclimation. a–f, Density of methylcytosines from the CpG context are shown for selected genes: trpm2 (a), pctp (b), cidea (c), gab1 (d), igf2 (e) and ddx6 (f). Red rectangles represent differentially methylated regions. Genomic locations are indicated below the density plot. Gene models are shown for the corresponding coordinates.
The paper's conclusion:
In conclusion, our study indicates that the epigenome is altered following exposure to increased temperatures via DNA methylation of specific loci. Although our results are consistent with transgenerational epigenetic effects, we cannot exclude a role for developmental epigenetic effects during the gamete and embryonic stage because eggs experienced the parental conditions until hatching. To conclusively demonstrate transgenerational epigenetic inheritance, future experiments should test if differential methylation and gene expression is retained when fish exposed transgenerationally to high temperature are returned to ambient control conditions in both parental and offspring generations. Nevertheless, we identified 193 DMGs that correlate to aerobic performance, of which many play key roles in metabolic homeostasis, insulin sensitivity and improved oxygen delivery, thus suggesting that these are the core genes associated with physical acclimation to heat stress across generations. Our study shows that exposure to higher temperatures associated with climate change causes genome-wide changes in DNA methylation, demonstrating that epigenetic regulation is possible in a coral reef fish facing a warming ocean, and that DNA methylation could play a role in transgenerational acclimation.
In my position as an advocate of nuclear energy, I often hear all kinds of idiotic remarks about mutations. Of course, radiation can and does cause mutations, but so do many other things, including, apparently doing exactly what we're doing about climate change, which is, um, nothing at all.
Have a pleasant day tomorrow.



