On the Road to an HIV Vaccine: Stimulating B Cells to Recognize HIV-1 V3 Glycans.
The paper I'll discuss in this post is this one: Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques (Nussenzweig et al Nature, Published On Line May 2019.
Recently I remarked in this space that when I was a stupid kid, I had a poor appreciation of the beauty and complexity of biologic molecules built from simple building blocks, i.e. peptides and proteins from amino acids, polynucleotides from nucleic acids, and, the topic of this paper, sugars.
Sugar chemistry has been difficult - for me at least - because it is kind of hard to represent on paper. For example, there are 16 "aldohexoses" - glucose being the most familiar - all of which have exactly the same formula, C6H12O6, differing only in their arrangements in space, i.e. their stereochemistry. Moreover, as a practical matter, each of these exist as a pair of "anomers" in which the stereochemistry at a single site shifts chemically, usually with two forms existing simultaneously.
Consider the three most common biological sugars:
Mannose:

Galactose:

Glucose:

Then consider that there are four forms of D-galactose:

...and since there are two forms of galactose which are mirror images of each other, eight kinds of galactose overall.
I was relieved of a small portion of my stupidity some years back when I stumbled upon a wonderful monograph, to which I've referred before in this space, The Sugar Code which has a heartwarming encouraging excerpt in the introduction stating that (I paraphrase): "No, your not as stupid as you think you are, sugar chemistry is hard"
The point of The Sugar Code is that this complexity of sugars, especially when they are linked together, particularly utilizing different sugars in one of two possible linkage systems, represents a remarkable opportunity to store information, an ability that living systems exploit extensively.
Modern advances in technology have made possible to interrogate these structures in hopes of understanding the signals they represent; recently I have been discussing this and other related topics in molecular biology with the major manufacturers of high resolution mass specs, and I'm amazed, totally amazed, at how remarkable this technology actually is.
An important tool for signaling in the immune system are called "glycans" which are an array of polymeric sugars and aminosugars that attach themselves to proteins, notably antibodies at N197, at asparagine (N-glycans) on the ? amide nitrogen, and serine (O-Glycans). The former are easier to analyze than the latter, although great strides have been made for the latter.
This brings me to the paper I cited at the beginning of the post. Signaling in the immune system is often highly involved in glycan signalling, and the text makes clear that antibodies evolve from one another in order to fight pathogenic infections with viruses, particularly those which are retroviruses, the mechanism for transcription in these offering many opportunities for error, and thus evolution.
In the language of the paper's introduction:
Single-cell antibody cloning from human donors who are infected with HIV-1 revealed that broadly neutralizing antibodies (bNAbs) have undergone unusually extensive somatic mutations1,2,3,4. Moreover, the high degree of somatic mutations is essential for binding to the native HIV-1 envelope (Env) spike and for the neutralizing activity of bNAbs5. The accumulation of large numbers of mutations suggests that bNAbs evolve in response to iterative rounds of somatic hypermutation and selection in germinal centres6. Studies in humans revealed that this occurs in response to viral escape variants that arise from antibody pressure4. Together, these observations suggest that vaccination to elicit bNAbs requires a series of sequential immunogens starting with an immunogen that induces the expansion7 of B lymphocytes expressing appropriate germline precursors8.
Sequential immunization to guide the development of bNAbs was demonstrated in genetically modified mice that carry inferred germline precursors of human bNAbs8,9. However, the priming immunogens that were used to initiate the response failed to activate and expand B cells that expressed inferred bNAb precursors in animals with polyclonal antibody repertoires. Thus, a goal of HIV-1 vaccine development has been to design immunogens that recruit B cells that express bNAb precursors into germinalcentre reactions in animals with polyclonal repertoires.
The authors have generated a B-Cell priming immunogen, which they call RC1, and explain in what might seem jargon to the uninitiated:
Here we describe RC1, an immunogen designed to recruit and expand diverse V3-glycan-specific B cells by improving accessibility of the V3-glycan patch epitope, which includes a group of high-mannose and complex-type N-glycans that surround V3 (Asn residues in gp120: N133, N137, N156, N295, N301, N332, N339, N385 and N392)15. bNAbs that target this site, including PGT12116, 10-107417 and BG1818, reach through these glycans using elongated heavy-chain complementarity-determining region (CDR) 3 (CDRH3) loops and portions of light-chain CDRs 1 and 3 (CDRL1 and CDRL3) to contact the highly conserved GDIR (G324-D325-I326-R327) motif at the base of V319. Here we show that RC1 activates and expands a diverse group of B cells expressing antibodies that resemble human V3-glycan patch bNAb precursors in mice, rabbits and rhesus macaques.
Some pictures from the text:
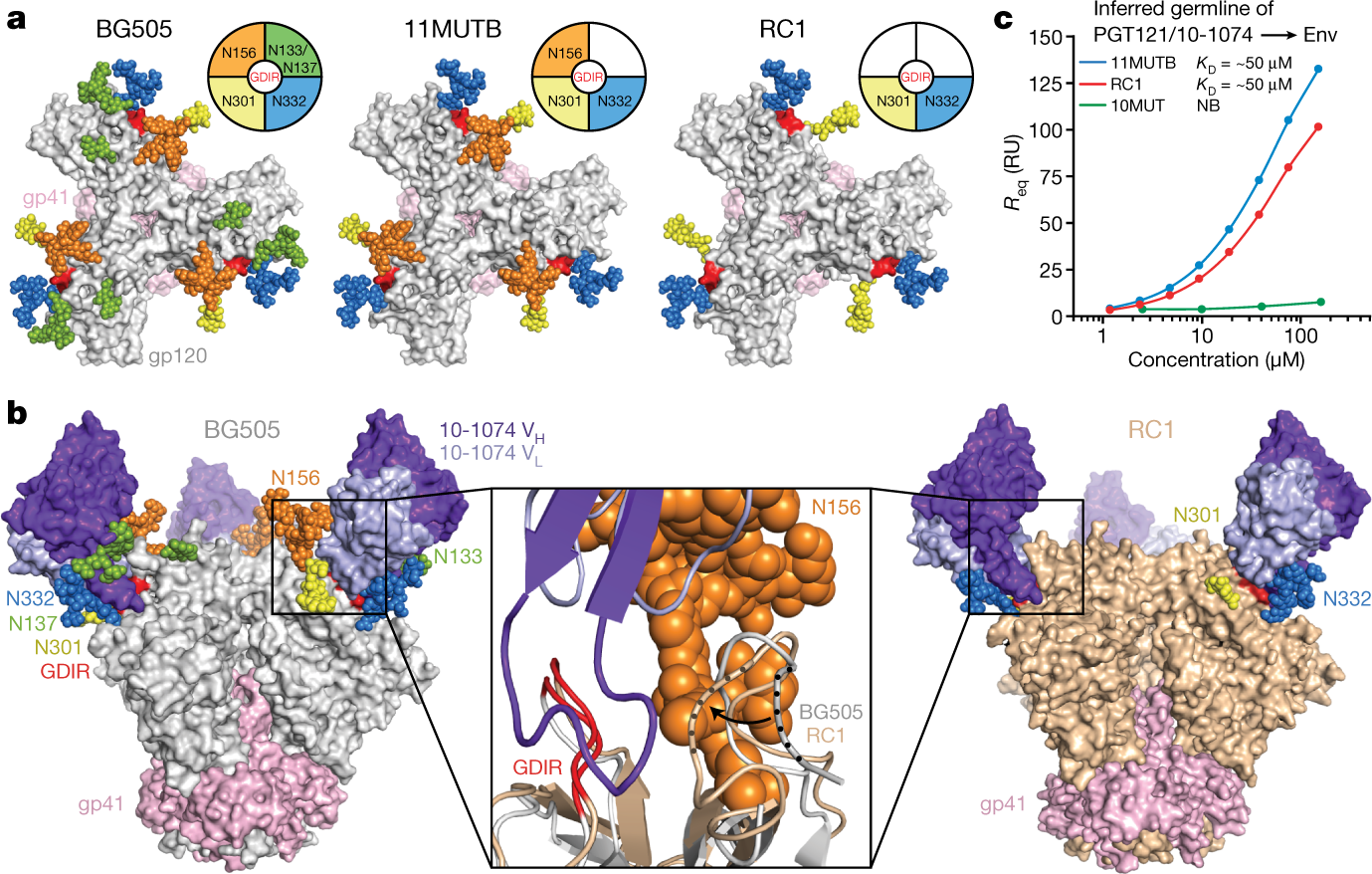
?as=webp
The caption:
a, N-glycans (coloured spheres) and GDIR motifs (red surfaces) mapped onto BG505 (Protein Data Bank (PDB) code 5T3Z) (N137 glycan from PDB 5FYL) in the top-down orientation. b, Left and right, side views of structures of BG505 and RC1 complexed with 10-1074 (glycan atoms are coloured spheres). Middle, superimposition of the boxed regions with protein in cartoon representations. Dark and light purple, 10-1074 VH and VL, respectively; red, GDIR; wheat, other portions of RC1; grey, BG505; orange spheres, N156 glycan. Regions of V1 showing displacement (gp120 residues 139–140) are indicated by dots and an arrow. c, Surface plasmon resonance (SPR) data for PGT121/10-1074 binding to Env trimers. NB, no binding above background; RU, response units. Representative plot from three independent experiments.
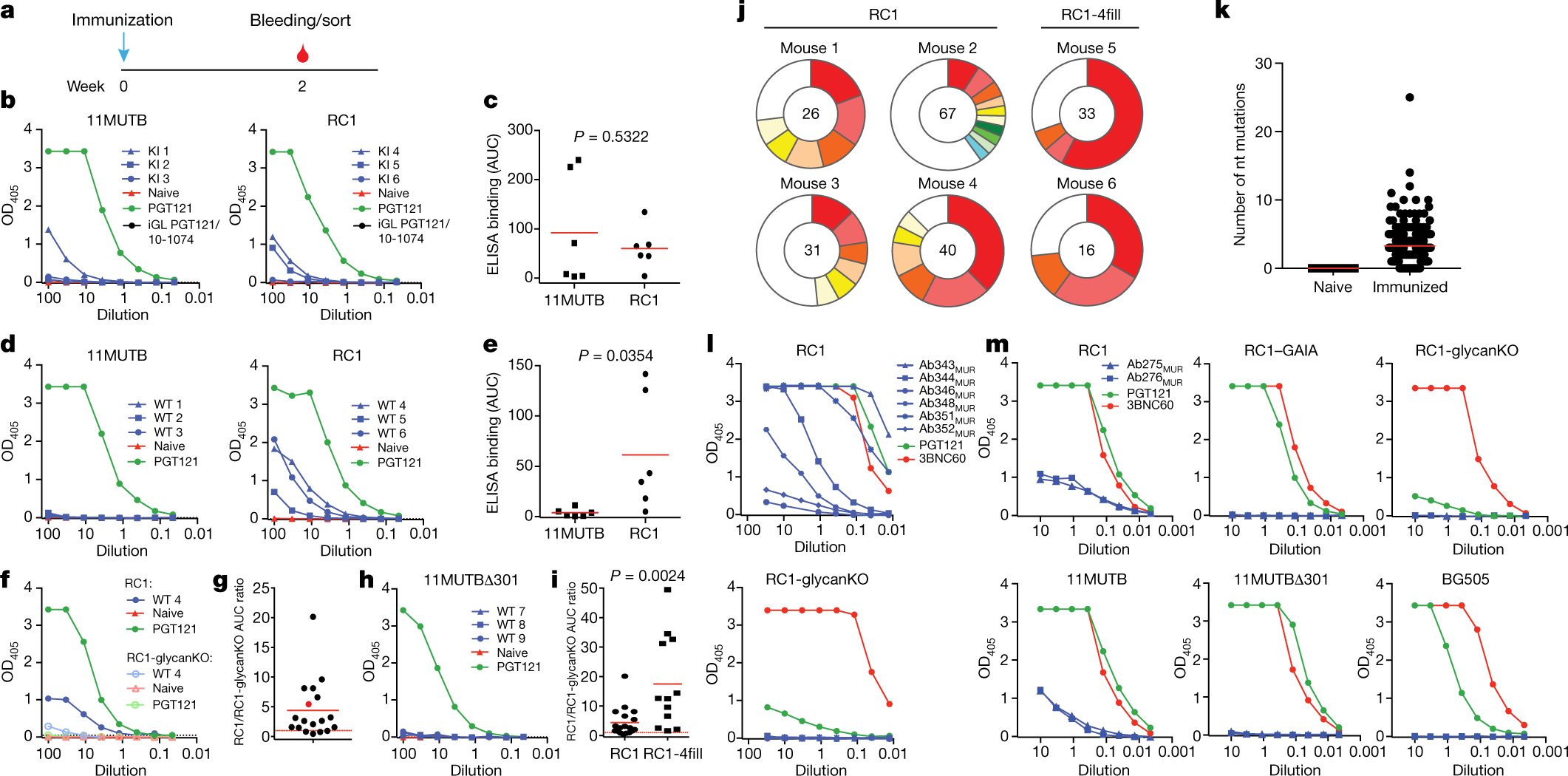
?as=webp
The caption:
a, Immunization protocol. b, d, f, h, Representative ELISAs showing serum binding to indicated immunogens. Controls include naive serum (red), purified PGT121 (green) and inferred germline (iGL) of PGT121/10-1074 (black). OD405, optical density at 405 nm. b, The inferred germline of PGT121 knock-in (KI) mice9. d, f, h, Wild-type (WT) mice. c, e, Area under the curve (AUC) for ELISAs in b and d, respectively, but combined results from two experiments using three or four mice each. Each dot represents serum from one mouse. f, Binding to RC1 and RC1-glycanKO. g, Ratio of the AUC for RC1 and RC1-glycanKO ELISAs from seven experiments with two or three mice immunized with RC1. Red dot corresponds to mouse WT 4 in f. i, Ratio of the AUC for RC1 and RC1-glycanKO ELISAs for wild-type mice immunized with RC1 (seven experiments) or RC1-4fill (five experiments). j, Pie charts show clonal expansion of RC1-binding B cells in the germinal centre. Coloured slices are proportional to the number of clonal relatives. White indicates single IgVH sequences. The number of heavy chains analysed is indicated in the centre. k, IgH nucleotide (nt) mutations from naive and RC1 immunized mice in j. l, ELISA binding of representative monoclonal antibodies from RC1-immunized mice to RC1 and RC1-glycanKO. m, ELISA binding of Ab275MUR and Ab276MUR to indicated Env proteins. c, e, i, Unpaired t-tests. c, e, g, i, k, Data are mean and each dot is an individual mouse (c, e, g, i) or an individual sequence (k).
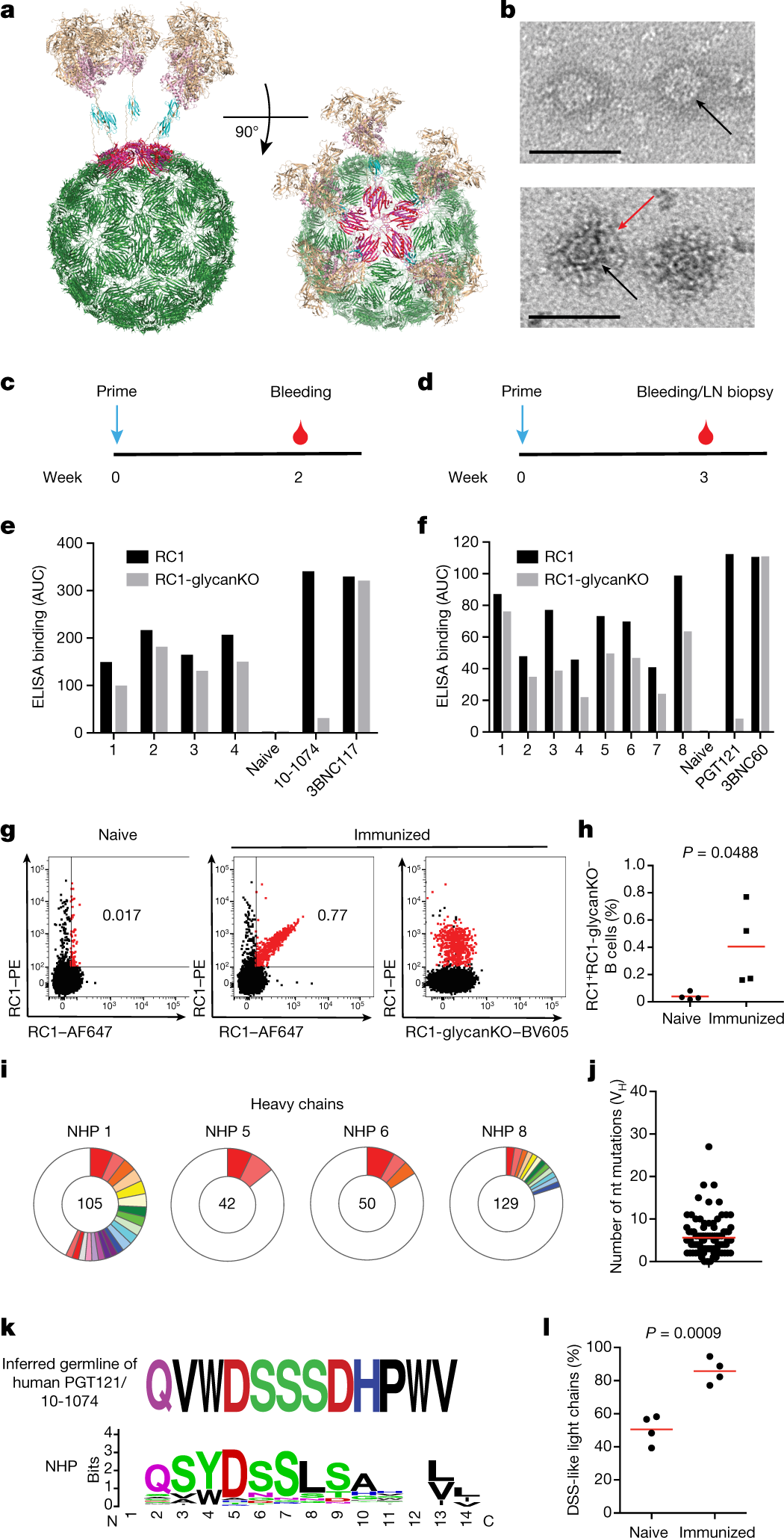
?as=webp
The caption:
a, Model of VLP-RC1-4fill: RC1-4fill (wheat and pink), SpyTag (gold), SpyCatcher (cyan) and bacteriophage AP205 (green). b, Negative-stain electron microscopy images comparing VLPs (top) and VLP-RC1 (bottom). Arrows indicate the VLP surface (black) and RC1 (red). Scale bars, 50 nm. Representative image from three independent experiments. c, d, Immunization protocols for rabbits (c) and non-human primates (d). LN, lymph node. e, f, AUC for ELISAs with serum from four rabbits (e) and eight non-human primates (f) primed with VLP-RC1-4fill against RC1 (black) and RC1-glycanKO (grey). g, h, Flow cytometry plots showing frequency of B cells in the germinal centre that bind to RC1 but not to RC1-glycanKO. g, Representative flow cytometry plots. h, Quantification. n = 4 naive and n = 4 immunized non-human primates. i, Pie charts showing clonal expansion of RC1-binding B cells in the germinal centre (see legend in Fig. 2j). j, IgVH mutations for the sequences of clones shown in i (Supplementary Table 3). k, Logo plots comparing CDRL3 of inferred germline of PGT121/10-1074 and all IgL from B cells in the germinal centre shown in i. l, Fraction of CDRL3 sequences from i that show a DSS-like motif. h, l, Unpaired t-test. h, j, l, Data are mean and individual values.
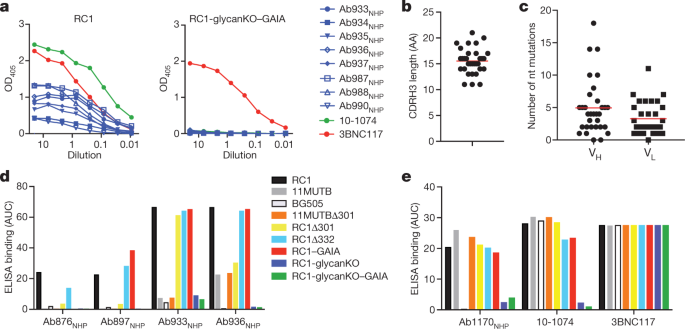
?as=webp
The caption:
a, ELISA binding of representative macaque monoclonal antibodies to RC1 and RC1-glycanKO–GAIA. b, CDRH3 length of 32 V3-glycan-patch-specific monoclonal antibodies. AA, amino acids. c, Nucleotide mutations in IgVH and IgVL of 32 V3-glycan-patch-specific monoclonal antibodies. d, e, AUCs for ELISA binding of monoclonal antibodies to indicated proteins. Each AUC value corresponds to one ELISA curve. b, c, Data are mean and each dot is an individual sequence.
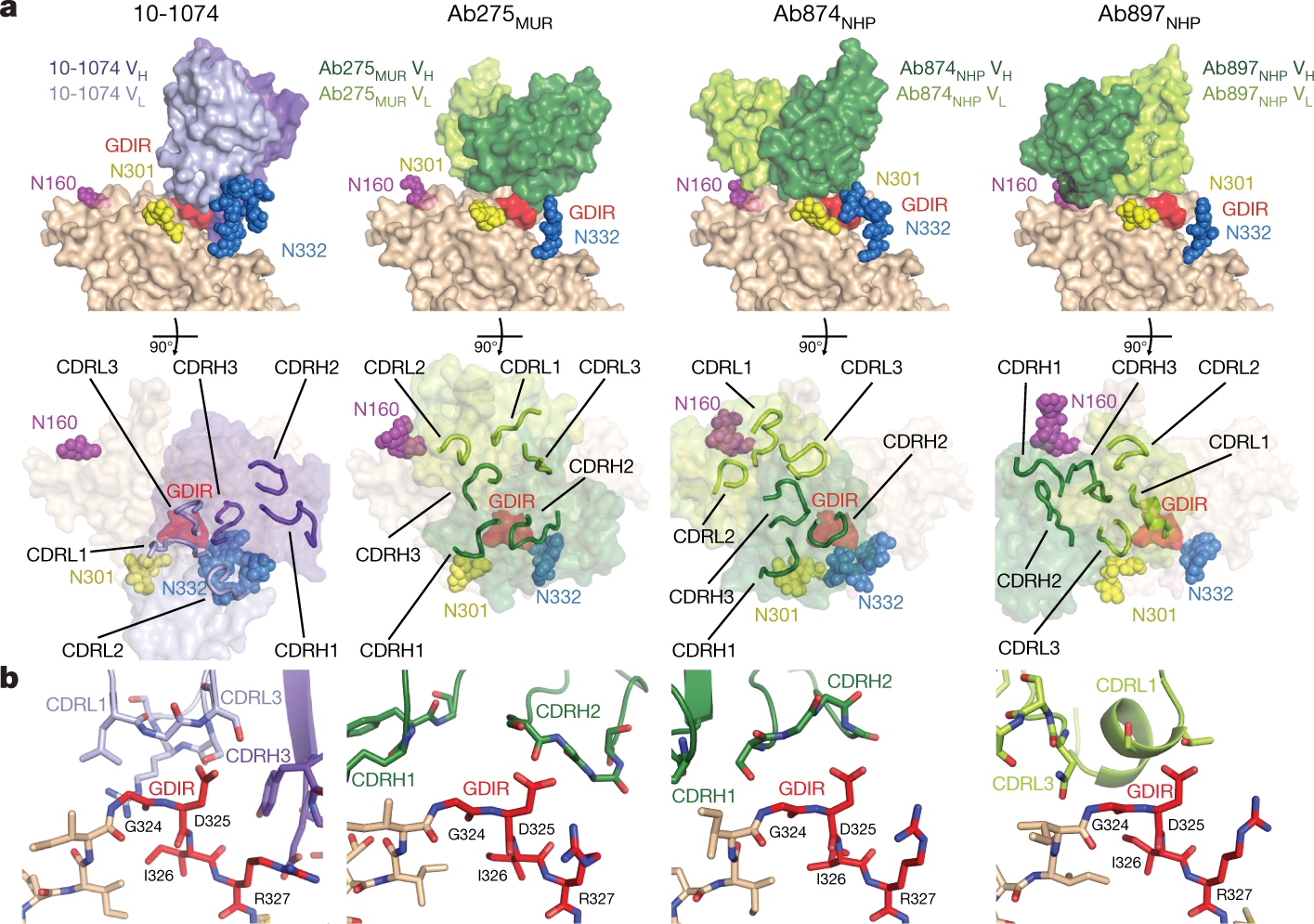
?as=webp
The caption:
a, Top, VH–VL domains of 10-1074 and elicited antibodies bound to one protomer of RC1 (GDIR residues are red; glycans are coloured spheres). Bottom, antibody combining sites (CDRs shown as loops) mapped onto gp120 (glycans as coloured spheres; GDIR in red). b, Comparisons of interactions of the GDIR motif with 10-1074 and elicited antibodies (colours as in a).
Some comments from the conclusion:
HIV-1 bNAbs develop in infected humans by sequential rounds of somatic mutation in response to a rapidly evolving pathogen4. Vaccination with a series of related antigens can reproduce this progression of events in genetically engineered mice that carry supraphysiological numbers of B lymphocytes that express the inferred germline precursors of bNAbs9. An important goal of HIV-1 vaccine design is to develop immunogens that initiate this response in organisms with polyclonal immune systems and then reproduce these responses in humans...
...The principles we used to produce RC1 did not take affinity for a germline B cell receptor into account. Instead, RC1 was designed to increase the number of bNAb progenitors that compete for entry into germinal centres by making the antigenic target site more available and facilitating binding to electrostatically neutral inferred germline precursors25. In addition, VLP-RC1-4fill incorporates the idea that masking competing off-target epitopes26,29 by addition of glycans27 and tethering the bottom of the trimer to a VLP minimizes competition for entry into the germinal centre.
RC1 differs from other HIV-1 vaccine candidates in that it induces B cells that express antibodies against a targeted epitope to undergo clonal expansion in germinal centres in animals with a fully polyclonal B cell repertoire...
...Notably, biochemical and structural results showed that antibodies with distinct mechanisms of targeting the V3-glycan patch were elicited by RC1, increasing the probability that one or more might develop breadth and potency after boosting9. Thus, VLP-RC1-4fill is a suitable candidate immunogen for further evaluation in sequential vaccination strategies to elicit bNAbs.
The development of vaccines has allowed for the total elimination of what was once a widespread fatal disease. The rise of and celebration of ignorance by the ignorant has made this goal less accessible than it was in former times, but with that caveat aside this is some very beautiful molecular biology.
Have a nice weekend.




 ?as=webp
?as=webp
 ?as=webp
?as=webp
 ?as=webp
?as=webp
 ?as=webp
?as=webp
 ?as=webp
?as=webp