NNadir
NNadir's JournalA Convergent Synthesis Scheme for Overcoming Virginiamycin Antibiotic Resistance.
The paper I'll discuss in this post is this one: Synthetic group A streptogramin antibiotics that overcome Vat resistance (Seipel et al., Nature (2020). https://doi.org/10.1038/s41586-020-2761-3)
Now that we are experiencing a global pandemic, we are more familiar with the state of affairs that existed for most of human history; deadly infectious agents routinely killed people in huge numbers, with the result that life expectancy was close to 1/2 of what it is today. My grandmother for instance, died in her early 40's from a bacterial infection - my mother was 11 years old - that today might have easily been cured with penicillin, or any other of a number of antibiotics in our arsenal.
However, since pathogenic bacteria have short doubling times, with many generations passing in a single day, there is ample opportunity for them to rapidly evolve resistance to those drugs, thus making them ultimately useless.
There is a big economic problem with anti-infectious medications and that is that they cure diseases. This is very different than is the case with, say, a blood pressure medicine that manages but doesn't cure the disease. The innovator company can collect sales for as long as they can keep its patent life going. Our economic system, given the extremely high cost and extremely risky nature of investing in the discovery of new drugs, does not select for curative agents (unless the disease is so widespread that sales will be enormous even if people are cured).
Worse from an economic standpoint is that if a drug can address a resistant strain that has evolved resistance to existing common antibiotics - well trained doctors will not write scrips for it unless all other medications have failed. This is responsible medicine: It follows that a drug that cures diseases that cannot be cured by "ordinary" means, will not sell.
When I was a kid, I briefly managed a combinatorial chemistry lab for a company that claimed - as marketing - that it was making tools that would enable the discovery of fifty drugs per year. The chemical libraries that I and my team made for this effort, under the direction of the chief scientific officer, didn't develop any drugs. Part way along in this effort, I figured out why this was inevitable, but nobody wanted to hear what I had to say, and I quit and found another job. (No, it was not as bad as Theranos; they were not making stuff up. It was just that the scientific assumptions did not hold experimental water.)
The paper I've cited above is kind of "chemical library" - like, a kind of better steered combinatorial chemistry. Don't give up the ship as they say. It is far superior in focus to what my company was doing 30 odd years ago.
From the introduction:
Streptogramin antibiotics comprise two structurally distinct groups (A and B)3 (Extended Data Fig. 1a) that act synergistically to achieve bactericidal activity in many organisms7 by inhibiting the bacterial ribosome8. Group A antibiotics bind to the peptidyl transferase centre (PTC) and increase affinity for the group B component in the adjacent nascent peptide exit tunnel9. Resistance to the A component mediates high-level resistance to the combination, whereas resistance to the B component results in intermediate resistance10. Similar to other antibiotics that target the PTC, resistance to group A streptogramins can be mediated by the ATP-binding cassette F (ABC-F) family proteins that dislodge antibiotics11 or by Cfr methylases that methylate A2503 of the 23S rRNA to sterically block binding12. A specific resistance mechanism for group A streptogramins is deactivation by virginiamycin acetyltransferases (Vats)2. These proteins acetylate the C14 alcohol, resulting in steric interference and disruption of a crucial hydrogen bond. The combination of vat(A) and vgb(A) genes (which deactivate the B component) is the most clinically relevant streptogramin-resistance genotype in S. aureus in France, where streptogramins (under the trade name Pyostacine) are used orally for skin and soft tissue infections13,14 as well as bone and joint infections15. Semisynthesis has improved water solubility (for example, Synercid16) and increased potency (for example, NXL-10317), but methods to overcome resistance to the class have yet to be discovered. Fully synthetic routes to group A streptogramins have been previously developed18,19,20,21,22,23,24,25,26,27,28, but these routes have not been applied to the synthesis of new analogues. Here we report optimization of our initially reported route18 and its application to the synthesis of analogues designed to overcome streptogramin resistance.
Streptogramins are antibiotics whose carbons are arranged (on a molecular scale of course) in large rings, rings much larger than the 5, 6, 7 and 8 member rings that are fairly common in chemistry.
The basic structure is suggested by this combinatorial chemistry synthetic scheme, Figure 1:
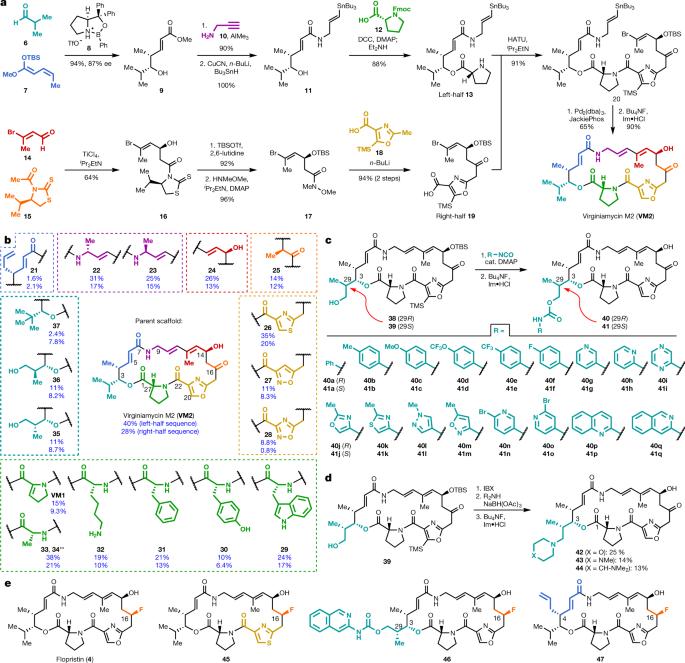
The caption:
In antibiotic talk, "MIC" is called the "minimum inhibitory concentration," the lower the number, the better the antibiotic:

The caption:
Years ago, the testing of potential drugs from chemical libraries basically depended on ligand binding assays. Thing have gotten far more sophisticated and now relies on actual measurement of protein-drug interactions:
To explore the structural basis for antimicrobial activity, we characterized several analogues bound to the E. coli ribosome using cryo-EM (Fig. 3c, d, Extended Data Figs. 5, 6). The PTC is highly conserved across pathogenic species of bacteria, and the E. coli ribosome is an appropriate model for group A streptogramin binding in both Gram-negative and Gram-positive organisms9. The 2.6-Å structure of analogue 47 bound to the ribosome clearly reveals the position of the C4-allyl extension, which projects towards the streptogramin B binding site and makes contacts with A2062, U2585 and U2586 (Extended Data Fig. 5). This extension also adopts a less strained conformation when ribosome-bound than when VatA-bound (calculated ?2.3 kcal mol?1) (Extended Data Fig. 7, Extended Data Table 2). This difference, along with protein conformational changes (Fig. 3b), could contribute to the observed differences in acetylation rates between 4 and 47. In the presence of VS1, the C4 extension adopts a strained conformation similar to its conformation in VatA but is probably stabilized by hydrophobic interactions with the B component (Fig. 3d).
Ligand strain may also have a role in the efficacy of 46. Predicted low-energy conformations of 46 position the arylcarbamate extension directly over the macrocycle (Extended Data Fig. 7); however, the structures of 46 bound to the ribosome in the presence or absence of VS1 (Fig. 3b, Extended Data Fig. 5) showed density for the extension in the P-site. The isoquinoline portion of the extension sits between A2602 and C2452, without making specific contacts with either. The proximity of C29 to U2585 may explain the difference in activity between the two diastereomeric series at this position (40a-q and 41a-q) (Fig. 1c).
A cartoon about the binding:
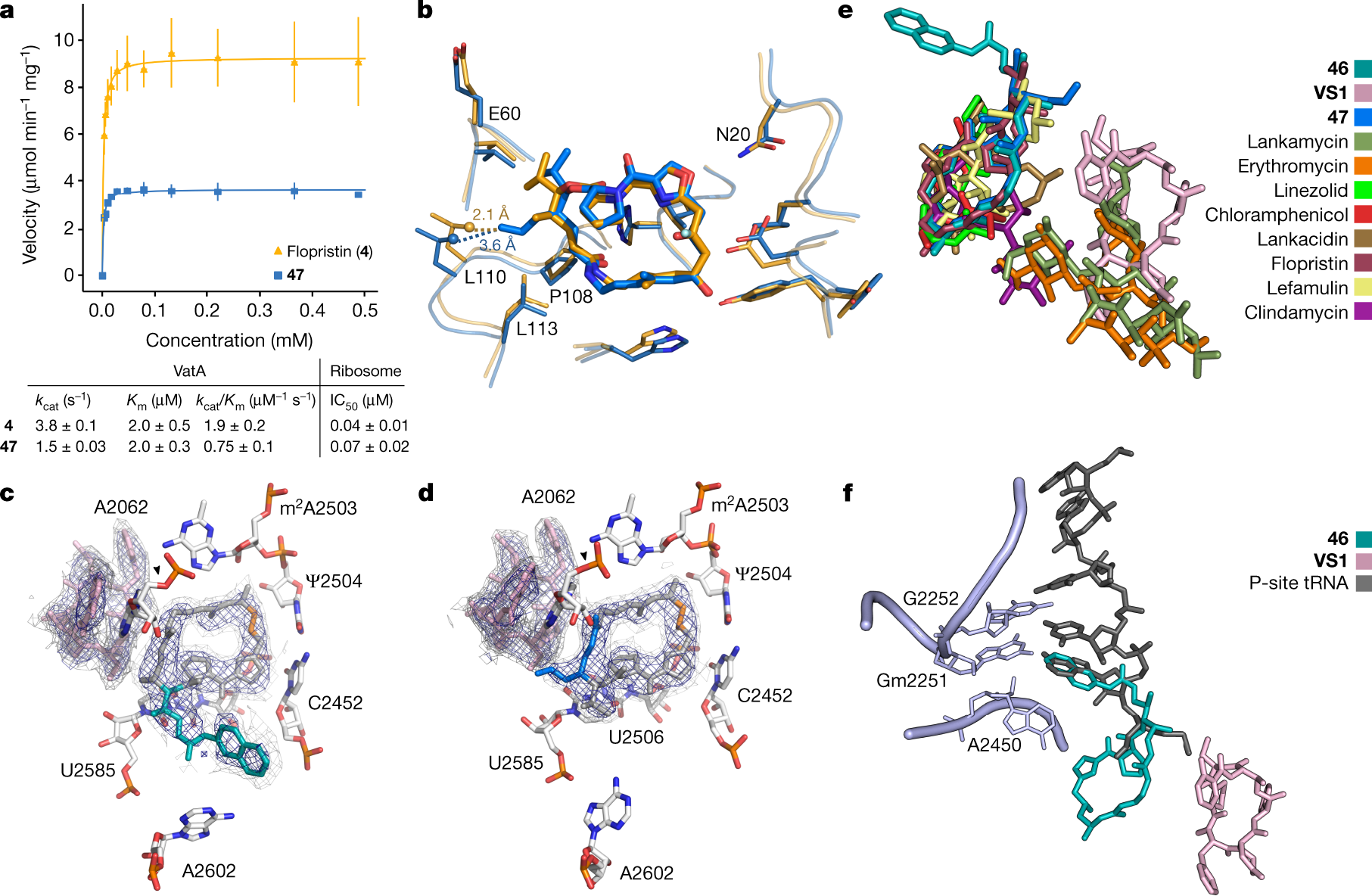
The caption:
Despite all this nonsense in the government, a true Confederacy of Dunces, to steal a line from John Kennedy Toole, scientists are still working to save our pathetic little butts from things other than Covid.
This is a cute little paper (actually a very, very good paper) that evokes a certain sense of nostalgia in me.
Life is interesting as hell, and then you die.
Have a nice day tomorrow.
...til Birnam wood comes to Dunsinane...
Cindy McCain endorsed a Democrat.
All of Shakespeare's tragedies concluded with a bloody justice, the victims honored, the villains destroyed by their own hubris.
"Despair thy charm and let the angel thought still have served tell thee. Macduff was from his mother's womb untimely ripped..."
Untimely ripped. Shakespeare wrote about real life, and although much in the United States States has been lost and cannot be restored i feel very much as if we are in the last act.
My son will not vote in New Jersey.
He registered to vote in Pennsylvania, where he's a student, and where he lives with his girlfriend.
He says Pennsylvania matters more than New Jersey.
His girlfriend - also from New Jersey - did the same.
I don't think, in any case, that he'll be coming home again. He graduates in December, will probably stay there for graduate school, seems to be in love, and there's no chance, absolutely none, that New Jersey will vote for the idiot racist fascist.
Helplessly Hoping
For what it's worth...
Population genomics of the Viking world
The paper I'll discuss in this post is this one: Population genomics of the Viking world (Eske Willerslev et al., Nature volume 585, pages390–396(2020))
According to "23 and me" analysis of my sister's-in-law genome, my wife is 30% (or maybe 32.38712% - I forget) French, which is news to me. I always kind of thought she was an American, and I have spent a lot of time in France among French people, I don't think my wife is French at all, or knows about how to be French, or wants to know about how to be French.
It gets a little silly in my view. These genetic ancestry companies, by the way, don't make any money by telling you about your remote relation to Anne Bolelyn's cousin's wife's brother's grandfather's adopted sister's great uncle's wife's bastard son. They make money by selling genetic distribution data to pharmaceutical companies, so they can figure out which diseases and syndromes are likely to be most profitable to manage and/or (god forbid) cure.
Nevertheless, there probably is some value in population genetics, and one area of active research is in physical anthropology, which can, if interpreted properly, surprise us by showing the commonality of our humanity, along with the beauty of diversity.
I came across this paper yesterday in desultory wandering around the literature, and it caught my eye because...because...well, honestly, I have no idea why it caught my eye but it seems to have done so. Following the references therein helped me make fun of one of my former intellectual conceits - which has now been considered to be vaguely (or maybe even grossly) racist - with respect to the disappearance of the Greenland Norse.
The main author listed in the reference next to the link - the authorship is quite a crowd - Eske Willerslev, is a pioneer in population genetics based anthropology, and according to his web page, is a world famous scientist and adventurer. Until yesterday, I never heard of him, but I'm a somewhat limited human being. For example, I know next to nothing about the Kardashian family except that one of them is apparently married to a famous African American Trumper, although I have no idea why the Trumper in question is famous.
Anyway, the abstract is available at the link above. From the introduction:
To explore the genomic history of the Viking Age, we shotgun-sequenced DNA extracted from 442 human remains from archaeological sites dating from the Bronze Age (about 2400 BC) to the Early Modern period (about AD 1600) (Fig. 1, Extended Data Fig. 1). The data from these ancient individuals were analysed together with published data from 3,855 present-day individuals across two reference panels (Supplementary Note 6), and data from 1,118 ancient individuals (Supplementary Table 3).
Here is a picture of some of the participants in this study, none of whom apparently gave "informed consent" to participate in the study, although there seems to have been no complaints from these subjects or their families about their uninformed lack of consent, which is probably a little less of ethical concern than say, the case of Henrietta Lacks, because the case of Henrietta Lacks involves some very questionable historical sociological implications:

The caption:
Examples of a few archaeological Viking Age sites and samples used in this study. a, Salme II ship burial site of the Early Viking Age, excavated in present-day Estonia: schematic of skeletons (top left) and aerial images of skeletons (top right, and bottom). b, Ridgeway Hill mass grave dated to the tenth or eleventh century AD, located on the crest of Ridgeway Hill near Weymouth, on the south coast of England (reproduced with permission from Dorset County Council/Oxford Archaeology). Around 50 predominantly young adult male individuals were excavated. c, The site of Balladoole, around AD 900, a Viking was buried in an oak ship at Balladoole (Arbory) in the south east of the Isle of Man. d, Viking Age archaeological site in Varnhem, in Skara municipality (Sweden). Schematic map of the church foundation (left) and the excavated graves (red markings) at the early Christian cemetery in Varnhem; foundations of the Viking Age stone church in Varnhem (middle) and the remains of a 182-cm-long male individual (no. 17) buried in a lime stone coffin close to the church foundations (right).
By the way, the supplementary files, which are probably open sourced, have fascinating details about all the dead people participating in the study. The files are 178 pages long, if you have some time to kill, and are found here: Supplementary information
The text continues:
The Viking Age Scandinavian individuals of our study fall broadly within the diversity of ancient European individuals from the Bronze Age and later (Fig. 2, Extended Data Figs. 2, 3, Supplementary Note 8), but with subtle differences among the groups that indicate complex fine-scale structure. For example, many Viking Age individuals from the island of Gotland cluster with Bronze Age individuals from the Baltic region, which indicates mobility across the Baltic Sea (Fig. 2, Extended Data Fig. 3). Using f4-statistics to contrast genetic affinities with steppe pastoralists and Neolithic farmers, we find that Viking Age individuals from Norway are distributed in a manner similar to that of earlier Iron Age individuals, whereas many Viking Age individuals from Sweden and Denmark show a greater affinity to Neolithic farmers from Anatolia (Extended Data Fig. 4a). Using the qpAdm program, we find that the majority of groups can be modelled as three-way mixtures of hunter-gatherer, farmer and steppe-related ancestry. The three-way model was rejected for some groups from Sweden, Norway and the Baltic region, which could be fit using four-way models that additionally included either Caucasus hunter-gatherer or East-Asian-related ancestry (Extended Data Figs. 4b, c)—the latter of which is consistent with previously documented gene flow from Siberia5,6,7.
Anatolia is the peninsula now wholly contained by the nation of Turkey, which is currently ruled by a Trump type, apparently. Irrespective of that fact, it's somewhat surprising to find that some Vikings were Turks.
The Viking World according to the authors, Fig. 1: Overview of the Viking Age genomic dataset:

The caption:
The next graphic apparently involves mapping from one real coordinate space to another (2 dimensional) coordinate space as a means of to determine "similarity" graphically. In one case, it's called "MDS" which is an abbreviation for multi-dimensional scaling and another, it's a "UMAP." I was frankly unfamiliar with these terms, which involve some nice mathematics involved in the area of statistics. Most of the statistics I know comes from taking courses in analytical chemistry, and some from osmosis over the years; but I've never actually taken a formal course in statistics, and certainly not in statistical similarity determinations in genomics. Poking around the internet showed me that apparently a UMAP is better than a tSNE, but I'm going in only so far as to say I've sort of, more or less, well, "in a fashion" have been there...
Anyway, similarity mappings, Fig. 2: Genetic structure of Viking Age samples:

The caption:
The authors continue:
The living and the dead, Fig. 3: Genetic structure and diversity of ancient samples.:


The caption:
And finally - trust me, Anne Boleyn is in here somewhere - Fig. 4: Spatiotemporal patterns of Viking and non-Viking ancestry in Europe during the Iron Age, Early Viking Age and Viking Age:



The caption:
Well then...
The authors discuss the disappearance of the Norse settlements in Greenland, an area of the world that is now famously melting at some threat to all of humanity, at least coastal humanity:
From around AD 980 to 1440, southwest Greenland was settled by people of Scandinavian ancestry (probably from Iceland)28,29. The fate of these populations in Greenland remains debated, but probable causes of their disappearance are social or economic processes in Europe (for example, political relations within Scandinavia and changed trading systems) and natural processes, including climatic change29,30,31.
According to our data, the Greenland Norse populations were an admixture between Scandinavians (mostly from Norway) and individuals from the British Isles, similar to the first settlers of Iceland18. We see no evidence of long-term inbreeding in the genomes of Greenlandic Norse individuals, although we have only one high-coverage genome from the later period of occupation of the island (Supplementary Note 10, Supplementary Figs. 10.2, 10.3). This result could favour a relatively brief depopulation scenario, consistent with previous demographic models32 and archaeological findings. We also find no evidence of ancestry from other populations (Palaeo-Eskimo, Inuit or Native American) in the Greenlandic Norse genomes (Supplementary Fig. 9.4)
I always like to wonder through the references in interesting papers, and as such, I found my way to Reference 30, which is this one:
Cultural adaptation, compounding vulnerabilities and conjunctures in Norse Greenland (Dugmore et al., PNAS March 6, 2012 109 (10) 3658-3663)
At the end of the last century, and the beginning of this one, it was intellectually fashionable to embrace the ideas of Jared Diamond, who won the Pulitzer Prize for his book "Guns, Germs and Steel" which was followed by "Collapse: How Societies Choose to Fail or Succeed. I waxed romantic about the latter book, which I read, in a post over at DailyKos - where I was happily banned for reporting a scientific truth, albeit in a very crude way - in order to support the obsessive viewpoint, which is either amusing or dire, of which anyone familiar with my writings will be aware. A large section of the book is about the disappearance of the Greenland Norse, which Diamond attributes to a refusal to eat Salmon because of superstition.
The more recent criticism of Diamond is that his work is overwhelmingly Eurocentric, and quite possibly, racist, at least in a vague, or perhaps obvious, way.
Reference 30 in the paper under discussion in turn contains reference 34, which is to Diamond's "Collapse" about which Dugmore and his coauthors say the following:
I love that locution referring to a logical fallacy, "special pleading," by which the authors dismiss Diamond's contention in "Collapse."
It has a certain trenchant eloquence, don't you think?
If there is any silver lining on the cloud of the near destruction of the United States by agency of having an ignorant racist lead it, it is that old white guys like me have had to reflect on their thinking and their assumptions, to question their immunity from being more than a little racist themselves, irrespective of their previous habits of thinking about themselves. This, I think, is a good thing. One is not really alive without questioning oneself.
The paper that is the general topic of this post concludes like this:
... Some Viking Age Scandinavian locations are relatively homogeneous—particularly mid-Norway, Jutland and the Atlantic settlements. This contrasts with the strong genetic variation of populous coastal and southern trading communities such as in the islands of Gotland and Öland47,48,49. The high genetic heterogeneity in coastal communities implies increased population size...
...Finally, our findings show that Vikings were not simply a direct continuation of Scandinavian Iron Age groups. Instead, we observe gene flow from the south and east into Scandinavia, starting in the Iron Age and continuing throughout the duration of the Viking Age, from an increasing number of sources. Many Viking Age individuals—both within and outside Scandinavia—have high levels of non-Scandinavian ancestry, which suggests ongoing gene flow across Europe.
Perhaps, just perhaps, the "Scandinavians" are more human than Scandinavian.
I trust you are having a pleasant, safe, and healthy Sunday afternoon.
Have you ever woke up in the morning and contemplated the life of Niels Ryberg Finsen?
Me neither.
Niels Ryberg Finsen
If you worry though, that this might happen to you, I advise you not to read the paper in this week's issue of Nature, on the exploration genetic signatures of the Vikings, Population genomics of the Viking world.
You might end up thinking about Niels Ryberg Finsen, and I'm sure you have better things to do.
I probably did too, but this is how my day ended up, finding out who Niels Ryberg Finsen was, and I'm worried that tomorrow might begin in a similar way, contemplating something or another about the Faroe Islands. (I haven't eaten a sheep since I was a child.))
Life is fun and then you die.
H. Holden Thorpe: "This may be the most shameful moment in the history of U.S. science policy."
This week's editorial in the journal Science, written by the Editor in Chief of this prestigious scientific journal, comments on Trump's decision to lie to the American people about Covid-19, thus killing and severely injuring large numbers of them:
EDITORIAL: Trump lied about science (H. Holden Thorp Editor-in-Chief, Science Journals. Science 18 Sep 2020: Vol. 369, Issue 6510, pp. 1409)
Over the years, this page has commented on the scientific foibles of U.S. presidents. Inadequate action on climate change and environmental degradation during both Republican and Democratic administrations have been criticized frequently. Editorials have bemoaned endorsements by presidents on teaching intelligent design, creationism, and other antiscience in public schools. These matters are still important. But now, a U.S. president has deliberately lied about science in a way that was imminently dangerous to human health and directly led to widespread deaths of Americans.
This may be the most shameful moment in the history of U.S. science policy...
...Trump also knew that the virus could be deadly for young people. “It's not just old, older,” he told Woodward on 19 March. “Young people, too, plenty of young people.” Yet, he has insisted that schools and universities reopen and that college football should resume. He recently added to his advisory team Scott Atlas—a neuroradiologist with no expertise in epidemiology...
...Monuments in Washington, D.C., have chiseled into them words spoken by real leaders during crises. “Confidence,” said Franklin Roosevelt, “thrives on honesty, on honor, on the sacredness of obligations, on faithful protection and on unselfish performance.”
We can be thankful that science has embraced these words. Researchers are tirelessly developing vaccines and investigating the origins of the virus so that future pandemics may be prevented. Health care workers have braved exposure to treat COVID-19 patients and reduce the death rate; many of these frontline workers have become infected, and some have died in these acts of courage. These individuals embody Roosevelt's call to faithful protection and unselfish performance.
They have seen neither quality exhibited by their president and his coconspirators. Trump was not clueless, and he was not ignoring the briefings. Listen to his own words. Trump lied, plain and simple.
Scientists act as if they are above and removed from politics at their own risk, and as pointed out here, at a risk to science itself.
Profile Information
Gender: MaleCurrent location: New Jersey
Member since: 2002
Number of posts: 33,512