Science
Related: About this forumA genomic timescale for placental mammal evolution
The article I'll discuss briefly in this post is this one: Nicole M. Foley et al., A genomic timescale for placental mammal evolution Science 380, 365 (2023).
In my day to day life, I am often called upon to consider protein sequences, and one of the most striking things is how many proteins are conserved across a wide array of species. Genetic and protein sequencing are now readily available tools, a long term benefit driven during the Clinton administration, a great scientific legacy of wise government. (The availability of these tools played a major role in stabilizing the recent Covid epidemic.)
Paleogenetics is a relatively new science, now about two decades old, for tracing the evolution of life on earth and establishing firm relationships between species. That's what this paper is about, specifically the evolutionary relationships to the class of organisms to which human beings belong, placental mammals.
From the introduction:
Prior studies have produced conflicting results regarding the timing and sequence of interordinal and intraordinal cladogenesis. As many as five models of placental mammal diversification have been proposed (4, 5), each implying different degrees of causality between the K-Pg extinction event and ordinal diversification. Each model is supported with molecular analyses of different sequence matrices that have been heavily biased toward short, evolutionarily constrained protein-coding exons or ultraconserved noncoding sequences (6–10). Biased genomic sampling has hampered a full resolution of the placental mammal phylogeny and an understanding of the principal drivers of ordinal diversification.
Here, we report a comprehensive analysis of phylogenomic signals from investigations of multiple genomic character types assayed from a hierarchical alignment (HAL) of 241 placental mammal whole-genome assemblies (1, 11). The HAL samples all placental mammal orders and represents 62% of placental families. The process and data structure that generated the HAL provide a statistically vetted whole-genome assessment of synteny and sequence orthology, reducing the potential for phylogenetic reconstruction errors caused by ortholog misidentification observed in some previous studies (12). The resulting availability of per base estimates of genomic constraint (PhyloP scores) also allowed us to assess the impacts of natural selection on phylogenetic signal and enabled the rigorous application of coalescent approaches (13)...
The authors utilized the data from the Zoomonia Consortium (cf. Zoonomia Consortium. A comparative genomics multitool for scientific discovery and conservation. Nature 587, 240–245 (2020), a curated tool for the construction of a database of the complete genomes of many animal species.
My own use of protein sequences is rather pedestrian and nowhere near the kind of computational work performed by my wife's boss (whose work with which I am not really familiar, except from osmosis from my wife's "shop talk." ) The following text, which I find somewhat arcane although I get the basic idea, reflects, I think, this sort of thing:
We applied site pattern frequency–based coalescent methods implemented in the SVDquartets program to sample single-nucleotide polymorphisms (SNPs) spaced by a minimum of 1 kb to reduce the impacts of intralocus recombination and linkage. We estimated phylogenetic relationships for all species in the HAL alignment and for 65 taxon matrices that sample all ordinal lineages while minimizing missing data (table S1). We analyzed three versions of the 65-taxon alignment to mitigate the reference-bias of alignments that were extracted from the HAL (table S2): a human-referenced alignment (HRA), a dog-referenced alignment (DRA), and a root-referenced alignment (RRA) that was imputed from the inferred placental ancestor (1). Because of the absence of nonplacental outgroups in our alignment, the root position was assumed to be between Atlantogenata and Boreoeutheria (5) and remains an open question. To investigate the impact of selection, we also identified conserved, accelerated, and nearly neutral evolving SNPs from a distribution of HRA sites ranked by PhyloP conservation scores across the 241-species alignment (14).
Some graphics from the paper:
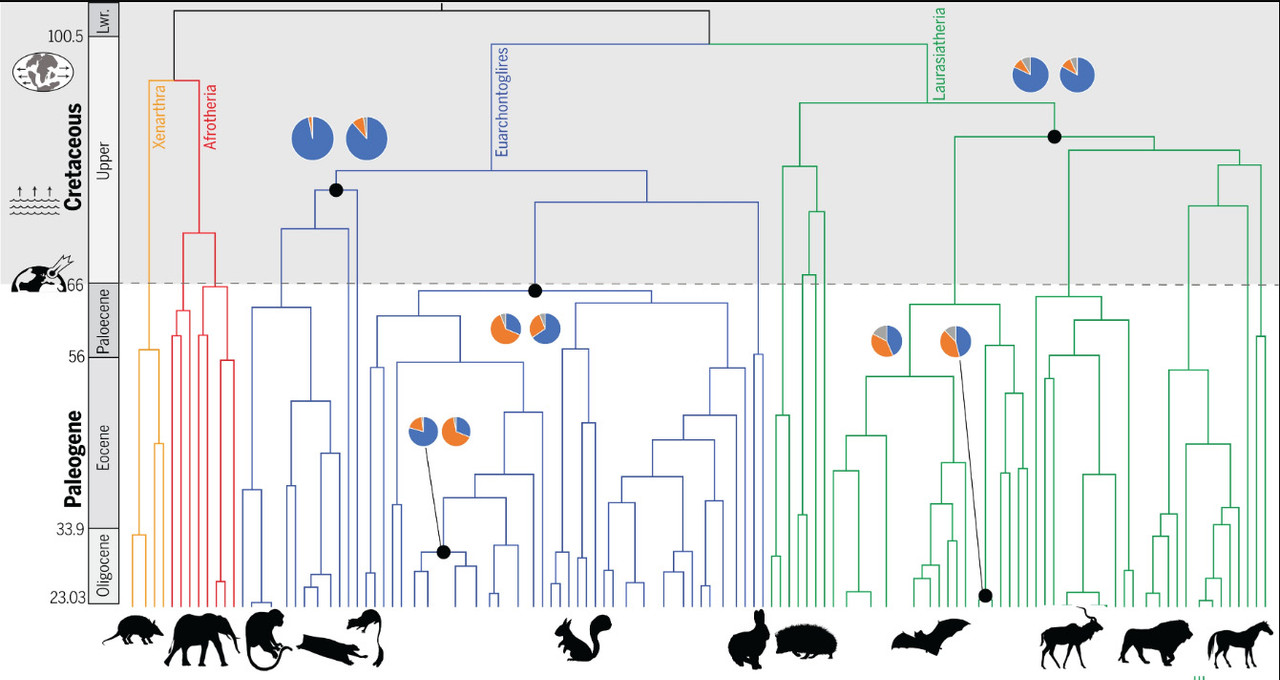
The caption:
Superordinal mammalian diversification took place in the Cretaceous during periods of continental fragmentation and sea level rise with little phylogenomic discordance (pie charts: left, autosomes; right, X chromosome), which is consistent with allopatric speciation. By contrast, the Paleogene hosted intraordinal diversification in the aftermath of the K-Pg mass extinction event, when clades exhibited higher phylogenomic discordance consistent with speciation with gene flow and incomplete lineage sorting.
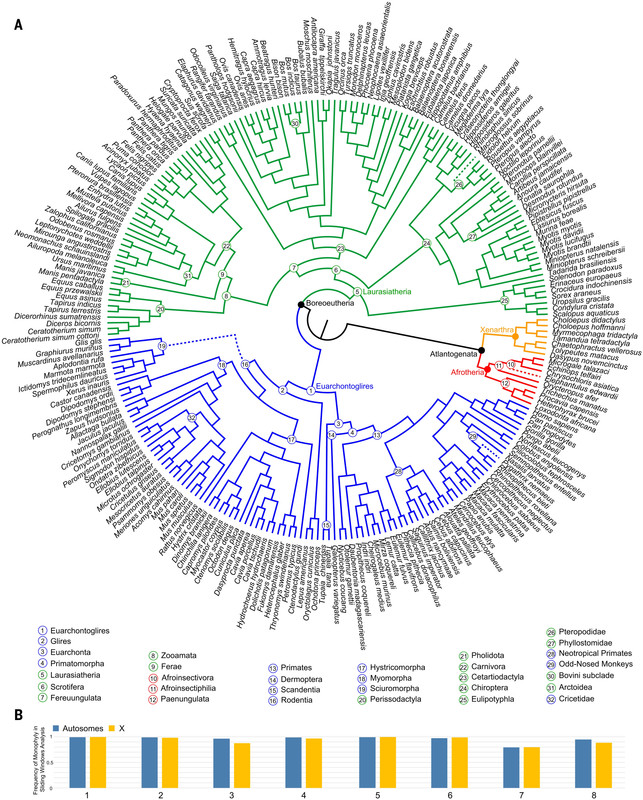
The caption:
(A) Fifty-percent Majority-rule consensus tree from a SVDquartets analysis of 411,110 genome-wide, nearly neutral sites from the human-referenced alignment of 241 species. Bootstrap support is 100% for all nodes. Superordinal clades are labeled and identified in four colors. Nodes corresponding to Boreoeutheria and Atlantogenata are indicated with black circles. (B) The frequency at which eight superordinal clades [numbered 1 to 8 in (A)] were recovered as monophyletic in 2164 window-based maximum likelihood trees from representative autosomes (Chr1, Chr21 and Chr22) and ChrX. Dotted lines indicate relationships that differ from the concatenated maximum likelihood analysis.
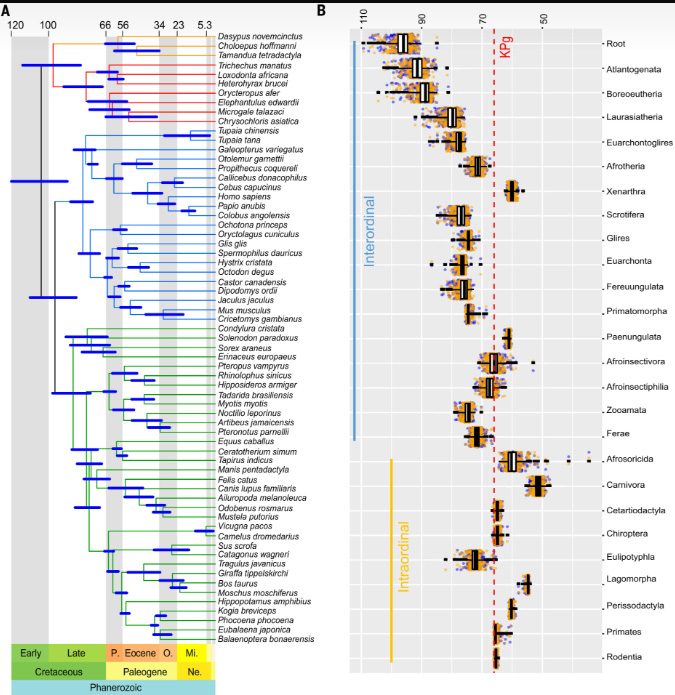
From the conclusion to the paper:
There are a number of concepts in this paper with which I lack routine familiarity, which means spending more time with it would be a worthy exercise.
Regrettably I will not have much time to go deeper into this work at present, but hope to return to it at some future time.
As we are living through a mass extinction event of our own creation, one hopes we can discover as many of these relationships as we can before they disappear.
Interesting.
Have a nice weekend.
Lemonwurst
(290 posts)The details are way over my head, but the context and graphics are so helpful for informing us regular folks about the depth of this kind of work. Fascinating.
4dog
(505 posts)but I’ll keep trying for a while.